
 |
||
05/07/10 |
|
|
The organisms I work with
How
I
ended up
working
with filamentous fungi and specifically Fusarium sp.
|
|
Fast growing | |
|
Relatively easy to genetically modify | |
|
Available Genome sequence | |
|
Crossing is possible | |
|
Genome wide expression data available (Affymetix platform) |
History and phylogenetics of the genus
The ascomycete genus Fusarium comprised over 1000 species before 1935, when Wollenweber & Reinkings laid the foundation for modern Fusarium taxonomy (Wollenweber & Reinking 1935). By comparing morphology and the reproductive characteristics of isolates on six different media, they reduced the number to 65 species, divided into 16 sections, based on the presence/absence and shape of microconidia, macroconidia and chlamydospores. The same year Raillo (1935) showed that some of the used characters were unsuitable for classification due to large variation within single isolates. With this new information Bilai (1950) reduced the number of species from 65 to 26, but this taxonomy did not win a wide acceptance. Snyder et al. (1957) further reduced the total number to 9 species by introducing a third name (cultivar) to the species name, indicating which host organism the subspecies showed pathogenity towards. The two classification schemes presented by Wollenweber & Reinking and Snyder et al. has been combined by Booth (1971) and later by Nelson et al. (1983) ultimately resulting in a 30 species system.
There is still debate on which characters are suitable for classification and therefore also on the number of species. The NCBI taxonomical database lists 47 Fusarium species, of which 37 are placed within the Gibberella fujikuroi species complex (http://www.ncbi.nlm.nih.gov/). To date only few species has been analysed with respect to rDNA sequence, as has been the standard for other genera (Samuels et al. 2001), (Tan & Niessen 2003). Future application of rDNA-based classification might offer a satisfying classification scheme for the taxonomy of the Fusarium genus, but until then, differences in morphological character-states will still be the deciding factor for identification and classification of Fusarium sp..
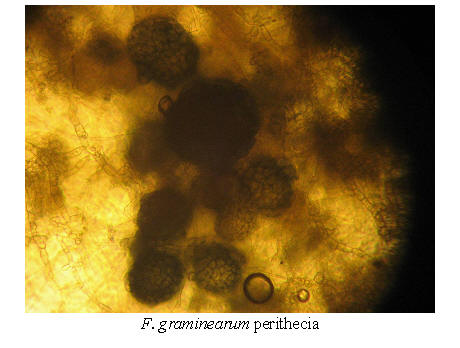
Fusarium graminearum, F. pseudograminearum and F. culmorum all belongs to the section "Discolor" in the Booth taxonomy. This section is characterized by thick-walled, distinctly septated macroconidia, with the ability to produce chlamydospores, but not microconidia (Booth 1971).
F. culmorum with no known teleomorphic state, was first described by Smith in 1884 and initially named Fusisporium culmorum, but renamed in 1895 to its present name also by Smith. The homothallic F. graminearum (teleomorph: Gibberella zeae) was first described in 1822 by Fries E. as Sphearia zeae, renamed in 1838 by Schwabe to its present name and linked to its teleomorphic state Gibberella zeae in 1936 by Petch (based on Booth 1971). In 1969, McKnight et al. showed that the F. graminearum species comprised two distinct pathogenic forms, one infecting maize and the other infecting cereals, for the following 33 years they were designated group 1 and 2, respectively. Upon closer examination by Aoki & O’Donnall the specie was split into to distinct species, group 2 remained F. graminearum (Gibberella zeae) while group 1 was renamed F. pseudograminearum (G. coronicola) (Aoki & O’Donnell 1999a). Aoki and O’Donnelll (1999b) also showed that F. pseudograminearum is heterothallic. F. graminearum is found in Europe, North and South America, Africa and Asia, while F. pseudograminearum currently only has been found in Asia and Australia. The relative narrow distribution of F. pseudograminearum is believed to be the result of its short history as a distinct specie, and that many scientists still classify them as F. graminearum.
Fusarium fujikuroi (teleomorph: Gibberella fujikuroi) is a plant pathogen that attacks rice, causing bakanae disease. The bakanae disease is characterized by elongation of the stem (hypertrophy) and seedling blight. The hypertrophy is due to the fungus production of gibberellic acid, a plant growth hormon. The thin elongated stems are typically not able to support the weight of the panicle, why they topple over, resulting in no production of edable grains (Kawaide 2006). The F. fujikuroi mycelium is red due to a production of bikaverin, a monomeric polyketide.
Fusarium proliferatum (Gibberella fujikuroi mating population D) is also a cosmopolite plant pathogen that attacks many important crops, such as rice (Desjardins et al. 1997), maize (Logrieco et al. 1995), date palm (Abdalla et al. 2000), asparagus (Elmer 1990) and onions (Stankovic et al. 2007). F. proliferatum has been reported to produce a broad range of mycotoxins, Fusaric acid (bacon et al. 1995), beaucericin (Logrieco et al. 1995), moniliformin (Marasas et al. 1984) and fumonisin B1 (Leslie et al. 1996).
For many of the Fusarium sp. sexual reproduction has not been observed and the genus are therefore classified as Deutoromycetes (fungi imperfecti). The fusaria species which has been linked to their respective teleomorphic stages are placed within the Ascomycetes. The lack of a teleomorphic stage can either reflect true reduction and loss of this stage (as seen in Penicillium sp.) or can be a result of inadequate information. Assignment of species to the Deutoromycete group is solely based on the absence of the sexual/teleomorphic stage (fungi perfecti) and the group does therefore not represent a true evolutionary group (Petersen 1998).
Members of the Deutoromycetes typically show parasexuality as a mean of genomic recombination. The parasexual cycle is dependent on anastomosis (fusion) of compatible haploid mycelia, resulting in the formation of heterokaryotic mycelia (Two types of nucleus in one cytoplasma) (Glass et al. 2003). The nuclei may in time fuse, giving rise to a diploid nucleus. This opens for the possibility of mitotic recombination between homologous chromosomes, and interchange of genetic material. The mycelium eventually becomes haploid again by random chromosome loss during successive mitotic divisions (Griffiths et al. 1999). This random chromosome loss also contribute to the formation of new genotypes (Sidhu 2002).
Dispersal and survival of the anamorphic state are ensured through vegetative (asexual) reproduction by formation of mitospores (known in Ascomycetes as conidia). Fusarium species features three types of conidia: microconidia, macroconidia and chlamydospores. The micro- and macroconidias are formed externally on hypha-like conidiophores, which categorizes Fusarium as a Hyphomycete. Chlamydospores are formed from normal hypha which undergoes increased growth and thickening of their cell wall (Deacon 1997).
The Fusarium genus contains some of the most important toxigenic plant pathogenic fungi. Their infection of important crops such as wheat, barley, oats, rice and maize pose a serious problem as infection leads to yield loss through lowered growth rate, reduction of grain size (Fusarium Head Blight/ tombstone/ scab (http://www.plantmanagementnetwork.org/pub/php/research/2003/fhb) and weakening of the straw (Foot rot) (Samson et al 2000), (Munkvold et al. 1998). In the US alone Fusarium outbreaks in the 1990's resulted in losses in the region of $3 billion (McMullen et al., 1997; Windels, 2000).
Bushnell et al. (2003) have described the infection mechanism of F. graminearum on wheat. Briefly summarized; The spores land on spikes and colonize the exterior of the plant. During anthesis the mycelium grow into the flowers and infect through weak points, such as stomata and anthers. Following the initial colonization, the mycelium easily spreads to the epidermis and parenchyma of the flower, stigmas and anthers. In some cases, the mycelium also resides between the kernel epidermis cell wall and cuticle. This growth pattern is believed to serve as a mechanism for dispersal of the fungi (Goswamin & Kistler 2004). The mycelium’s penetration is thought to be aided by excretion of hydrolytic enzymes (cutinases, celluloses, amylases and pectinases) (Jenczionka et al. 2003).
Overvintering is achieved either in the chlamydospore state or as a saprophyte on plant debris left in the field. Schmidt & Feistritzer showed that F. culmorum can remain viable for up to 2 years buried at a depth of 50 cm (according to Booth 1971). This ability poses a substantial threat to the following year’s crop. Counter-measures for preventing reinfection, includes crop rotation and deep burial of the previous crop before seeding, with crop rotation as the most efficient of the two [http://www.planteinfo.dk/cp/graphics/Name.asp?Language=da&TaskID =4&NameID=236].
Favorable conditions for perithecia and macroconidia formation results in the dispersal of the formed ascospores (sexually formed spores) and macroconidia (asexualspores) to plants. Ascospores and macroconidia are typically spread by wind, rain or insects (Parry et al. 1995).
Apart from field infections, F. culmorum is also known to course storage rot of sugar beet, potatoes and apples. Storage rot of cereals is typically a secondary effect of the primary field infection (Goswami & Kistler 2004).
Many Fusarium species
may under the right conditions turn pathogenic toward
animals
(invasive fusariosis). A successful invasion is dependant on depression of
the victim’s immune system, as found in cancer, AIDS, bone marrow transplant
patients or even in cases of a common cold (Beardall & Miller 1994).
Invasion of the blood stream is quite common (in this aspect Fusarium
sp. differs from other opportunistic fungal animal pathogens), but has
little effect on healthy individuals. As a testimony to this a large portion
of the general population has antibodies against Fusarium cell
surface polysaccharides (Notermans et al. 1988). F. solani, F.
oxysporum and F. moniliforme are the species most commonly
causing fusariosis.
Fusarium sp. are generally soil born, but surveys has shown that
Fusarium under certain conditions is more abundant in air samples than
Aspergillus and that 17 % of throat samples from otherwise healthy
people contain Fusarium sp.. Ocular infections by F. solani
has also been observed especially in contact lens users, as 14 % of all soft
contact lenses are infected with Fusarium sp. without being invasive.
Fusarium infections are typically treated with Amphotericin B,
natamycin and rifampicin (Nelson et al. 1994).
Fusarium graminearum, isolate PH-1, was sequenced in 2003 as a collaboration between the Broad Institute (MIT) and the International Gibberella zeae Genomics Consortium (IGGR). The sequence is public available at the Broad Institute home page (called FG1) and it has later been annotated by the Munich Information Center for protein sequences (MIPS) and is available at Fusarium graminearum Genome DataBase (FGDB) (called FG2). The annotation resulted in approximately 14.000 gene models based on EST data and gene prediction software (Calhoun annotation system, FGENESH and manual annotation (GENEMARK, GENEFINDER, GENSCAN)), the genes are distributed on four chromosomes with a total size of 36 Mb (Broad). Approximately 50 % of the genome encode genes, giving an average gene density of 1 gene pr. 3100 bp (Güldener et al. 2006). Over 50 % of the called genes do not show similarity to any previously characterized genes/proteins, showing that there still is a long way to go for functional analysis.
The development of the F. graminearum GeneChip (Affymetix platform) allowed for a more direct annotation of the included genes based on hybridization results from individual probes, this has resulted in a third gene set (FG3), which is accessible through both FGDB and the Broad Institute (Güldener et al. 2006). Expression data obtained from the Affymetix GeneChip is public accessible through www.plexdb.org.
The genomes of Fusarium oxysporum and Fusarium verticillioides (Gibberella moniliformis) were sequenced in 2005 and 2007, also by the Broad Institute, and are accessible through the "Fusarium Comparative Database". In 2007 Fusarium solani (Nectria haematococca) was sequenced by JGI (DOE Joint Genome Institute) and the gene calls are available through "JGI N. haematococca v2.0 Home".
The availability of four Fusarium genomes now opens for the possibility of comparative genomics and more efficient gene calling. Currently the four genomes have been annotated independently, however when doing comparative analysis it quickly becomes apparent that the gene calls for the four genomes do not always match, as extensive synteny between the genome sequences are not always reflected in the gene calls. A project for a simultaneous gene calling for three of the species have been initiated and received funding - and we all currently awaits the release of the new gene sets.
|
Abdalla, M. Y., Al-Rokibah, A., Moretti, A., & Mule`, G. (2000). Pathogenicity of toxigenic Fusarium proliferatum from date palm in Saudi Arabia. Plant Disease, 84, 321–324. | |
|
Aoki, T. and K. O’Donnell “Morphological and molecular characterization of F. pseudograminearum sp. nov., formerly recognized as the Group 1 population of F. graminearum”, Mycologia (1999a), Vol. 91, p. 597-609 | |
|
Aoki, T. and K. O’Donnell “Morphological characterization of Gibberella coronicola sp. nov., obtained through mating experiments of Fusarium pseudograminearum”, Mycoscience (1999b), Vol. 40, p. 443-453 | |
|
Bacon, C. W., Porter, J. K., Norred, W. P., & Leslie, J. F. (1996). Production of fusaric acid by Fusarium. Applied and Environmental Microbiology, 62, 4039–4043. | |
|
Beardall J.M and Miller J.D. “Diseases in humans with mycotoxins as possible causes” p. 487-539 in “Mycotoxins in grain. Compounds other than aflatoxin”, edited by J.D. Miller and H.L. Trenholm, Eagan Press, St. Paul, Minn, USA (1994) | |
|
Bilai V.I. “The Fusaria (biology and systematics)”, Akademia Naukwe Ukrinskii SSR, Kiev (1950), only the abstract in English is available | |
|
Booth C. “The Genus Fusarium” Commenwealth Mycological Institute, The Eastern Press Limited London (1971). | |
|
Bushnell W.R., Hazen B.E. and Pritsch C. “Histology and physiology of Fusarium head blight.”, “Fusarium Head Blight of Wheat and Barley” Ed. Leonard K.J. and Bushnell W.R., APS Press, St. Paul, p. 44-83, (2003) | |
|
Deacon J.M. “Modern Mycology, 3th edition” published by Blackwell Science, printed in Cambridge (UK) by the University Press (1997) | |
|
Desjardins, A. E., Plattner, R. D., & Nelson, P. E. (1997). Production of fumonisin B1 and moniliformin by Gibberella fujikuroi from rice from various geographic areas. Applied and Environmental Microbiology, 63, 1838– 1842. | |
|
Elmer, W. H. (1990). Fusarium proliferatum, as casual agent in Fusarium crown and root rot of asparagus. Plant Disease, 74, 938. | |
|
Glass N.L. and Kaneko I. “Fatal Attraction: Nonself Recognition and Heterokaryon Incompatibility in Filamentous Fungi”, Eukaryotic Cell (2003), Vol. 2, No. 1, pp. 1-8 | |
|
Goswami, R. S. and H. C. Kistler. 2004. Heading for disaster: Fusarium graminearum on cereal crops. Molecular Plant Pathology 5:515-525 | |
|
Griffiths A.J.F., Miller J.H., Suzuki D.T., Lewontin R.C. and Gelbart W.M. “Introduction to Genetic Analysis 7th edition” printed in New York for W. H. Freeman & Co. (1999) | |
|
Jenczionka N.J., Maier F., Lösch A.P. and Schäfer W. “Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal of the head-blight disease of wheat, are regulated by the MAP kinase gpmk1”, Current Genetics (2003), Vol. 43, p. 87-95 | |
|
Güldener U., Manhaupt G., Münsterkötter M., Haase D., Oesterheld M., Stümpflen V., Mewes H-W. and Adams G. (2006). FGDB: a comprehensive fungal genome resource on the plant pathogen Fusarium graminearum. Nucleic Acids Res. 34(Database issue): D456–D458. | |
|
Güldener U., Seong K.Y., Boddu J., Cho S., Trail F., Xu J.R., Adam G., Mewes H.W., Muehlbauer G.J. and Kistler H.C. (2006). Development of a Fusarium graminearum Affymetrix GeneChip for profiling fungal gene expression in vitro and in planta. Fungal Genet Biol. 43(5):316-25. Epub 2006 Mar 13. | |
|
Kawaide H. "Biochemical and molecular analyses of gibberellin biosynthesis in fungi", Biosci Biotechnol Biochem. (2006) Vol. 70, No. 3:583-90 | |
|
Leslie, J. F., Marasas, W. F. O., Shephard, G. S., Sydenham, E. W., Stockenstrom, S., & Thiel, P. G. (1996). Duckling toxicity and the production of fumonisin and moniliformin by isolates in the A and F mating populations of Gibberella fujikuroi (Fusarium moniliforme). Applied Environmental Microbiology, 62, 1182–1187. | |
|
Marasas, W. F. O., Nelson, P. E., & Toussoun, T. A. (1984). Toxigenic Fusarium species: Identity and mycotoxicology. University Park: Pennsylvania State University Press. | |
|
McKnight T. and Hart J. “Some field observations on crown rot disease of wheat caused by Fusarium graminearum”, Queensland Journal of Agricultural and Animal Science (1966), Vol. 23, p. 373-378 | |
|
Logrieco, A., Moretti, A., Ritieni, A., Bottalico, A., & Corda, P. (1995). Occurrence and toxigenicity of Fusarium proliferatum from preharvest maize ear rot, and associated mycotoxins, in Italy. Plant Disease, 79, 727–731. | |
|
McMullen,
M., R. Jones, and D. Gallenberg. 1997. Scab of wheat and barley: A
re-emerging disease of devastating impact. Plant Dis. 81:1340-1348 | |
|
Nelson P. E., Dignani M. C. and Anaissie E. J. “Taxonomy, Biology, and Clinical Aspects of Fusarium Species”, Clinical Microbiology Review (1994), Vol. 7, No. 4, p. 479-504 | |
|
Nelson, P.E.,
Toussoun, T.A., and Marasas, W.F.O., “Fusarium species” The Pennsylvania
University Press, USA (1983) | |
|
Parry D.W.,
Jenkinson P. and McLeod L. “Fusarium ear blight (scab) in small grains – a
review”, Plant Pathology (1995), Vol. 44, pp. 207-238 | |
|
Raillo A. “Diagnostic estimation of morphological and cultural characters of species in the genus Fusarium sp.”, Bull. Plant Prot II Leingrad (Phytopathol) (1935), Vol. 7, p. 1-100 | |
|
Samson R.A., Frisvad J.C. and Hoekstra E.S. “Introduction to Food- and Airborn fungi, sixth edition” printed by American Society Microbiology (2000) | |
|
Samuels G.J., Nirenberg H.I. and Seifert K.A. “Perithecial species of Fusarium” in “Fusarium - Paul E. Nelson Memorial Symposium” edited by Summerell B.A., Leslie J.F., Bachhouse D., Bryden W.L. and Burgess L.W. APS press, St. Paul (2001), USA | |
|
Sidhu G.S. “Mycotoxin genetics and gene clusters”, European Journal of Plant Pathology (2002), Vol. 108, p. 705-711 | |
|
Snyder W.C., Hansen H.N. and Oswald J.W. “Cultivars of the fungus Fusarium”, Journal of Madras University, B 27, p. 185-192, (1957) | |
|
Tan M.K. and Niessen L.M. “Analysis of rDNA ITS sequences to determine genetic relationships among, and provide a basis for simplified diagnosis of, Fusarium species causing crown rot and head blight of cereals”, Mycological research (2003), Vol. 107, No. 7, p. 811-21 | |
|
Stankovic, S., Levic, J., Petrovic, T., Logrieco, A. & Moretti, A. (2007) Pathogenicity and mycotoxin production by Fusarium proliferatum isolated from onion and garlic in Serbia. Eur J Plant Pathol, vol. 118, p. 165–172 | |
|
Windels, C. E. 2000. Economic and social impacts of Fusarium head blight: Changing farms and rural communities in the Northern Great Plains. Phytopathology 90:17-21 | |
|
Wollenweber
H.W. and Reinking O.A. “Die Fusarium, ihre Beschreivung, Schadwirkung und
Bekämpfung” Paul Parey, printed in Berlin (D) (1935) |
Dette sted blev sidst opdateret 05. July 2010